Principles of qPCR
Quantitative polymerase chain reaction (qPCR) is a molecular biological technique used to quantitatively analyze the target DNA sequence in a sample. The target DNA sequence is amplified by primers, and the number of PCR products is detected by fluorescent probe or SYBR Green I fluorescent dye method, thus achieving accurate quantification of the target sequence. qPCR technology has the advantages of fast, accurate, high sensitivity and high specificity, so it is widely used in gene expression analysis, mutation detection, pathogen detection and other fields.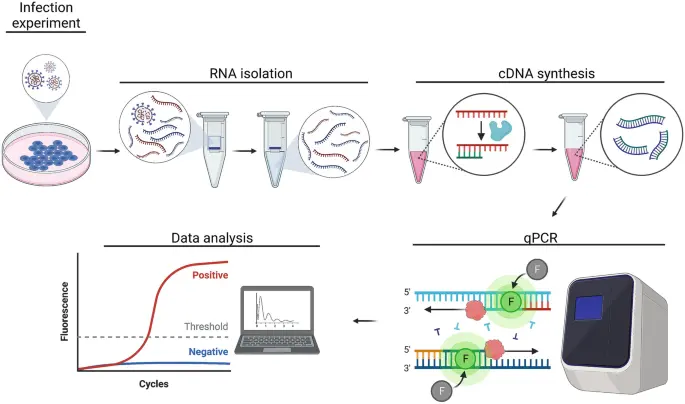
Materials and Instruments
Reagents: RNA extraction kit, reverse transcription reagent, qRT-PCR kit, cell inflammatory factor primer (probe method also needs to be equipped with probe), PBS, DEPC water, etc.Experimental materials: RNase-free centrifuge tube and eight-row PCR tube, RNase-free gun tip, disposable gloves, etc., to prevent RNA degradation and sample contamination.
Instruments and equipment: centrifuge, ultra-clean workbench, ultraviolet spectrophotometer, qRT-PCR instrument, etc.
Experimental Procedure
Cell culture: The cells to be tested are inoculated in a petri dish and the cells are treated specifically according to the purpose of the experiment. Before collecting cells, gently wash with PBS buffers 2-3 times to remove components in the medium that may interfere with subsequent experiments.RNA extraction: Total RNA is extracted from cells using an RNA extraction kit. RNA concentration and purity are measured by ultraviolet spectrophotometer, and usually A260/A280 values between 1.8-2.1 indicate good quality RNA samples. Since RNA is very unstable and easily degraded, it is recommended to perform a reverse transcription reaction as soon as possible after the assay is completed.
cDNA synthesis: According to the reverse transcription reagent instructions, it is usually divided into two steps: first, DNase digestion is used to remove genomic DNA, and then reverse transcription reaction is performed to synthesize cDNA. After this step is completed, the cDNA sample can be frozen at -80℃ for subsequent experiments.
qRT-PCR reaction: specific primers are designed and synthesized, cDNA is used as a template, and qRT-PCR amplification is performed with specific primers. After the reaction, the amplification curve and the melting curve were confirmed and analyzed according to the standard curve.
Data analysis: Fluorescence signal was detected by qRT-PCR instrument and Ct value was derived. Using β-actin as internal reference gene, the expression level of inflammatory cytokines was calculated by 2-△△CT method, and standardized and statistical analysis was performed by data analysis software.
Matters Needing Attention

RNA extraction: The quality and purity of RNA are critical for cDNA synthesis and qRT-PCR results. Therefore, it is necessary to use high-quality RNA extraction kits and operate in strict accordance with the instructions to avoid RNA degradation and contamination.
cDNA synthesis: Select the appropriate reverse transcriptase, primer and reaction conditions to ensure that the RNA is efficiently transcribed into cDNA. In particular, DNase digestion must be performed before reverse transcription to prevent DNA contamination from affecting subsequent detection.
Primers and fluorescent probe design: When detecting cellular inflammatory factors, specific primers and fluorescent probes need to be designed to ensure specific amplification of the target sequence and reduce the occurrence of false positives. Appropriate primers and probes can be designed using an online tool such as Primer3, or purchased directly from a supplier.
SYBR Green dye method: If the SYBR Green dye method is used, it is necessary to determine whether ROX dye needs to be added according to the model of PCR instrument used to correct for changes in the fluorescence signal.
Data analysis: The result analysis of qRT-PCR requires the help of professional software, data standardization and statistical verification. It is recommended to repeat the experiment several times to ensure the reliability of the results.
Sum up
To sum up, qPCR technology, with its unique advantages, plays an irreplaceable role in genome research and clinical diagnosis. However, to obtain accurate and reliable results, it is necessary to strictly follow the code of practice and ensure the high quality of the execution of each step. In addition, through reasonable design of experimental scheme and optimization of parameter Settings, further improve the specificity and sensitivity of detection, is the direction of future technology application and research. Mastering and using qPCR technology will bring greater value and prospect for scientific research and clinical application.
Next: Common Pipetting Errors and Prevention
Previous: Pulse Biological News about Pipette Tips

